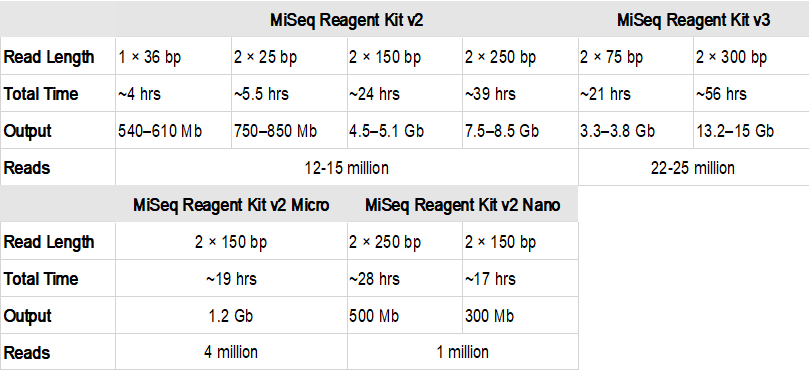
paired end sequencing read length
The library prep protocols are designed to. This size depends on the library.

Jacob Schreiber On Twitter There S A Second Even More Insidious Issue Here Read Length The Longer The Read Length The Longer The Fragment Lengths We Can See Here That All Of The
Requires the same amount.
. The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase the. Whether you align 100bp paired reads or a. The paired-end short read lengths are always 2 x 150bp 300bp.
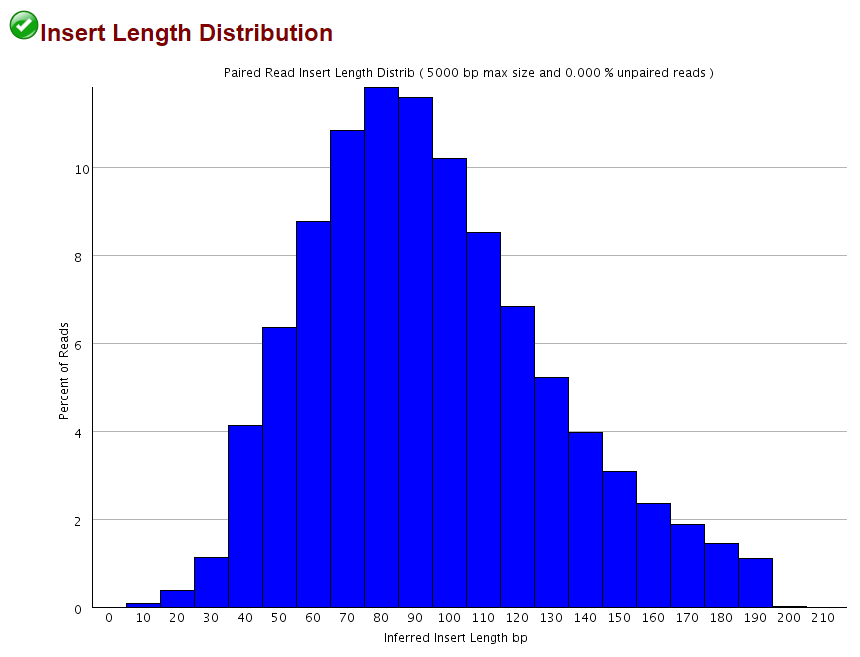
For example one read might consist of 50 base pairs 100 base pairs or more. A good choice for read length is closely tied to the insert size of the sequencing library ie how long the individual DNA fragments are that are sequenced. When you align them to the genome one read should align to the forward strand and the other should align to the reverse strand at a higher base pair position than the first one.
When using RNA-Seq to study gene expression read length is not a significant factor. Enter up to 20 characters or use manual mode if you need between 20 and 100 bp. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts.
What matters is read counts. We use an Illumina MiniSeq for our short-read sequencing runs. Since paired-end reads are more likely.
The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase the. Application of sequencing to RNA analysis RNA-Seq whole transcriptome SAGE expression analysis novel organism mining splice variants Search in titles only Search in RNA. Simple workflow allows generation of unique ranges of insert sizes.
To ensure sequencing quality of the Index Read do not exceed the supported read. During sequencing it is possible to specify the number of base pairs that are read at a time.
Short Read Sequencing Genomics Core Ecu

Rapid Low Cost Assembly Of The Drosophila Melanogaster Reference Genome Using Low Coverage Long Read Sequencing Biorxiv
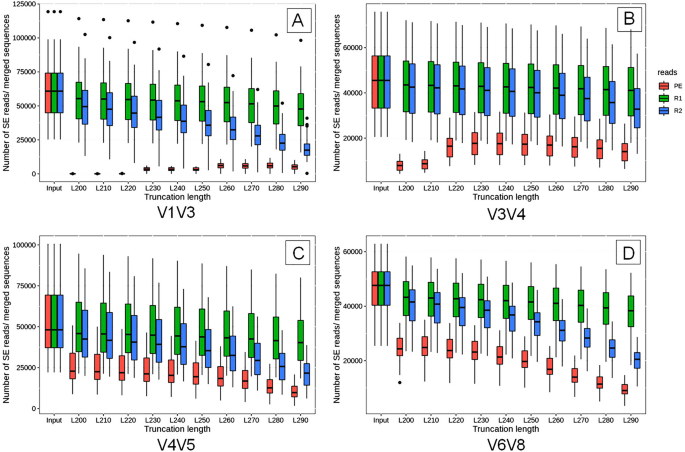
A Comprehensive Evaluation Of Single End Sequencing Data Analyses For Environmental Microbiome Research Springerlink
Rdp Tutorials Initial Processing Tool

Comparison Of Next Generation Sequencing Systems
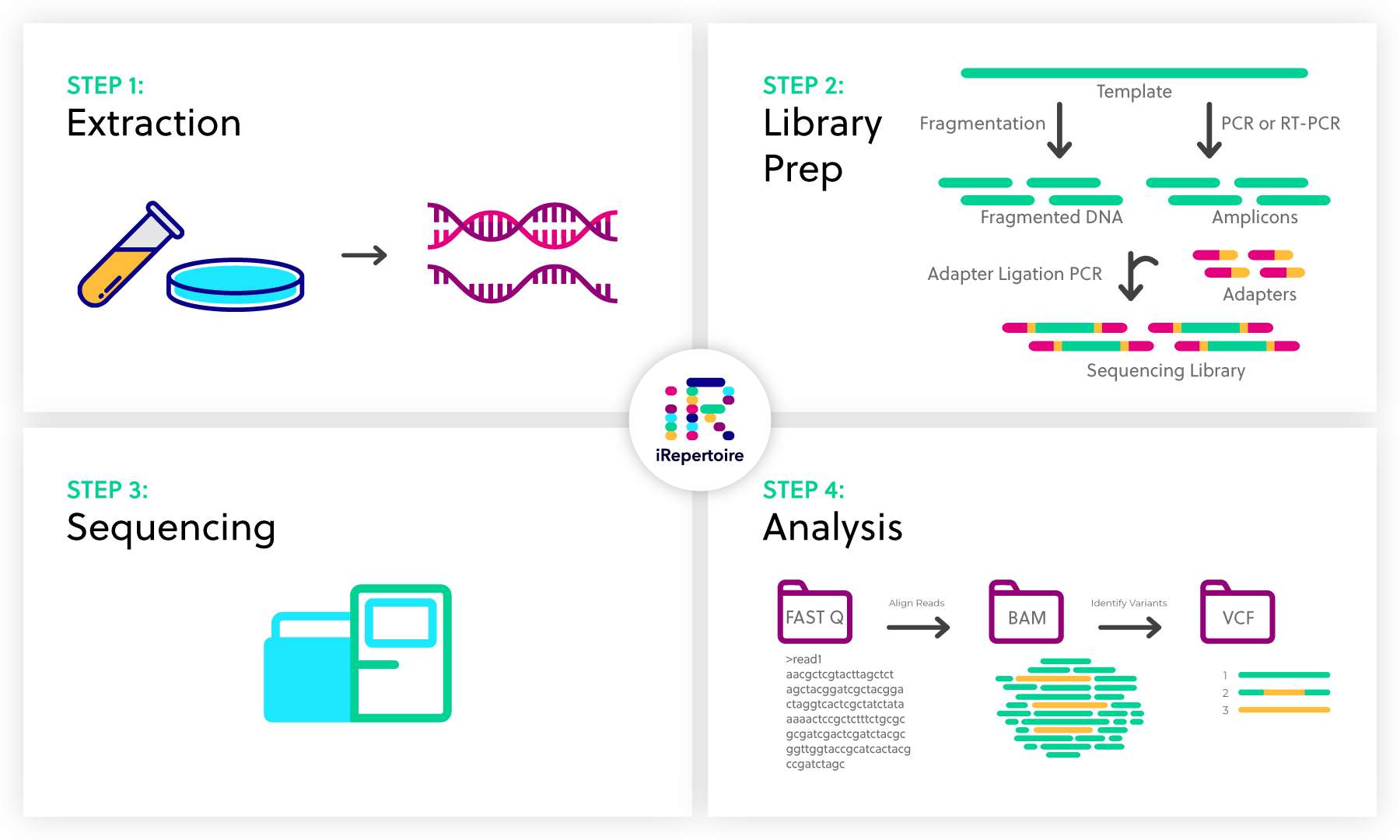
Next Generation Sequencing Ngs Overview Irepertoire Inc
Qc Fail Sequencing Mispriming In Pbat Libraries Causes Methylation Bias And Poor Mapping Efficiencies

Skewer A Fast And Accurate Adapter Trimmer For Next Generation Sequencing Paired End Reads Topic Of Research Paper In Biological Sciences Download Scholarly Article Pdf And Read For Free On Cyberleninka Open Science
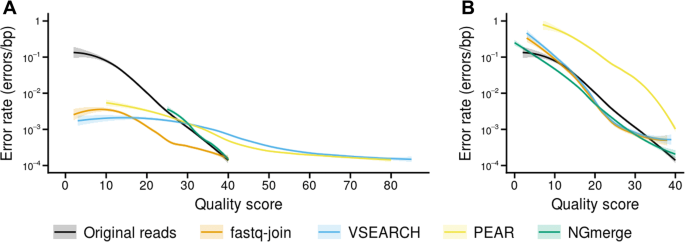
Ngmerge Merging Paired End Reads Via Novel Empirically Derived Models Of Sequencing Errors Bmc Bioinformatics Full Text
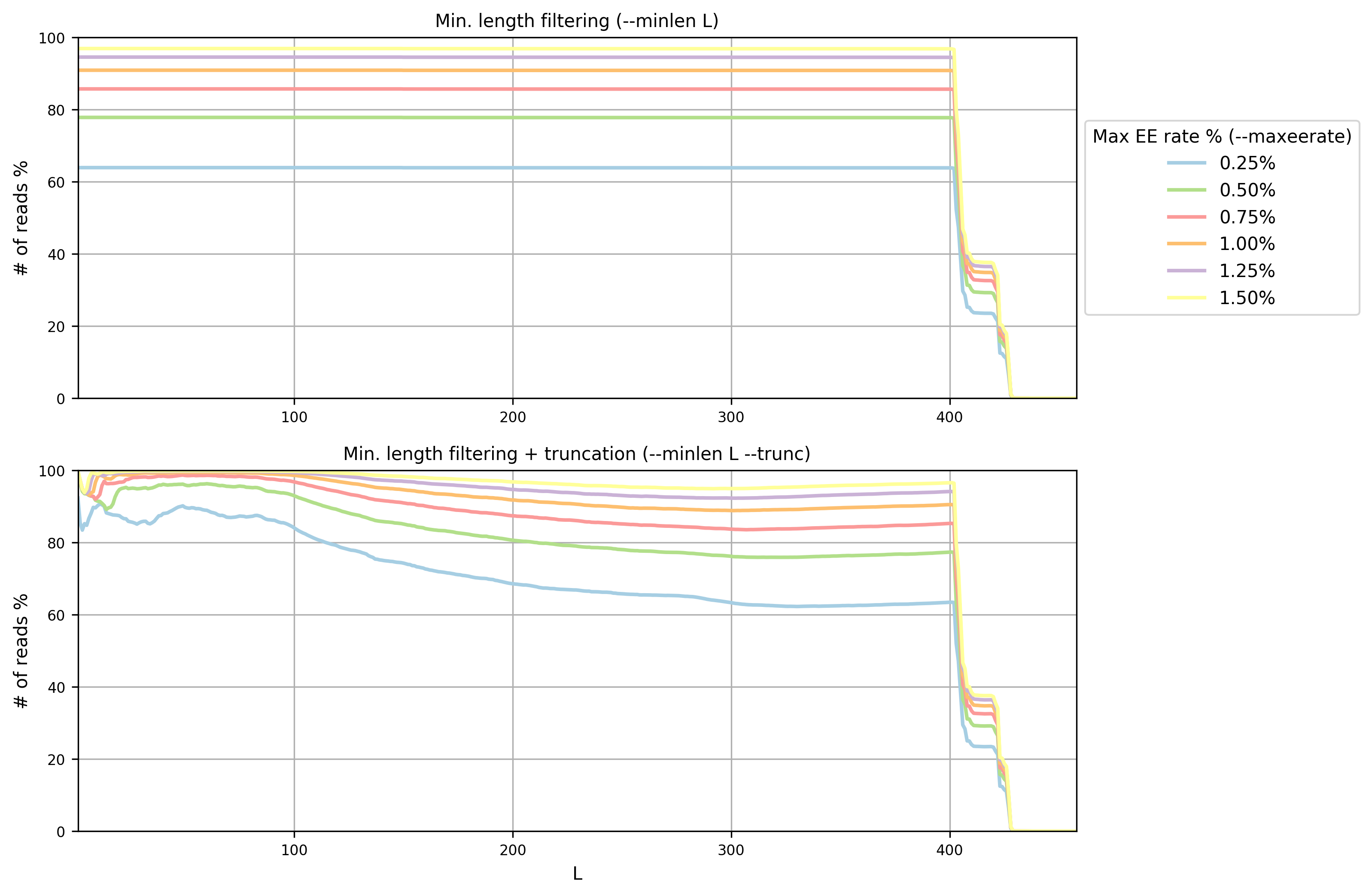
Paired End Sequencing 97 Otu Micca 1 7 0 Documentation

Schematic Representation Of Paired End Sequencing For Very Short Download Scientific Diagram
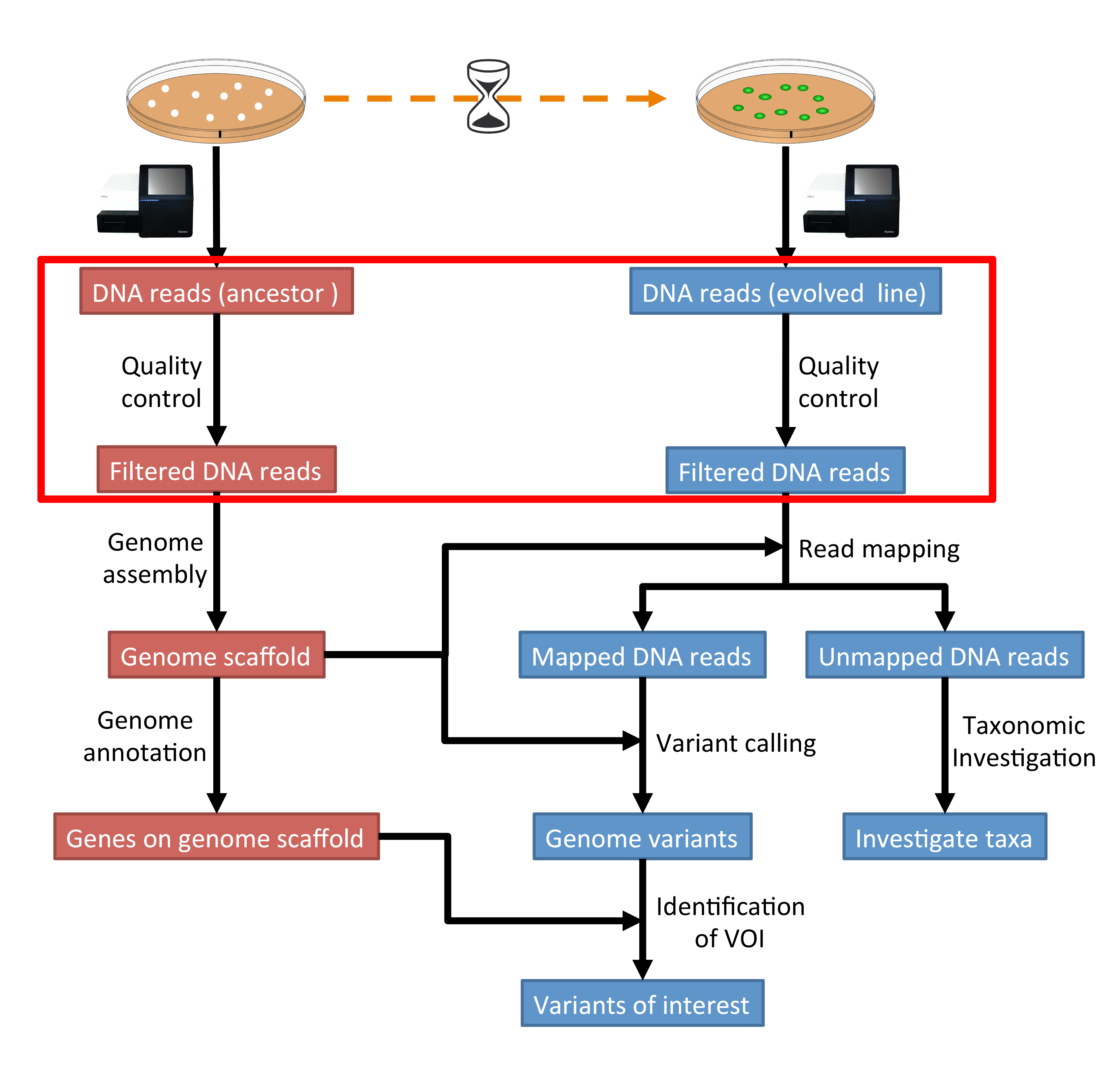
3 Quality Control Genomics Tutorial 2020 2 0 Documentation

An Overview Of Next Generation Sequencing Technology Networks
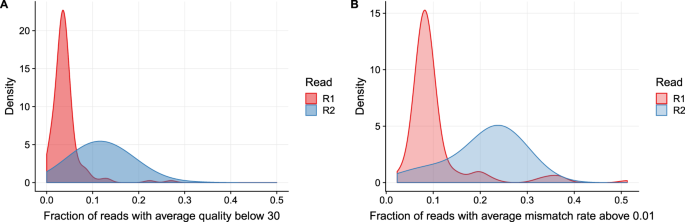
Long Fragments Achieve Lower Base Quality In Illumina Paired End Sequencing Scientific Reports
What 2 Indicates In 2 150 Bp 2 300 Bp Maximum Read Length From Illumina Website Http Www Illumina Com Systems Sequencing Platforms Html
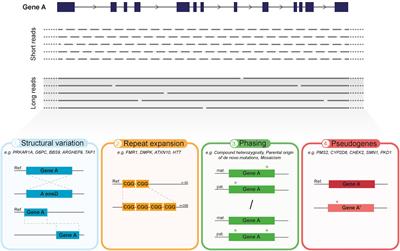
Frontiers Long Read Sequencing Emerging In Medical Genetics

